This page gives you an example that you can reproduce using the MyHits environment. Please follow the steps in "purple".
Starting from one single protein, you will do a PSI-BLAST on Swiss-Prot, select the proteins of interest, modify the multiple alignment with Jalview and send everything back to PSI-BLAST for another search cycle.
The process is similar for PFSEARCH and HMMER
Starting from one single protein, you will do a PSI-BLAST on Swiss-Prot, select the proteins of interest, modify the multiple alignment with Jalview and send everything back to PSI-BLAST for another search cycle.
The process is similar for PFSEARCH and HMMER
|
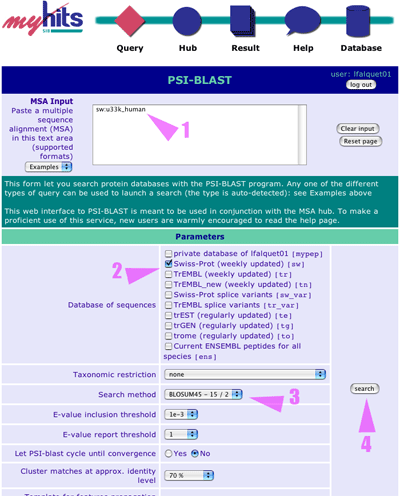
First use the Query menu and select PSI-BLAST on any page to access the PSI-BLAST form, then follow these steps: 1. Type the "sw:UBXN1_HUMAN" protein identifier in the input text area 2. Check the Swiss-Prot database 3. Select matrix "BLOSUM45" 4. Click on the search button |
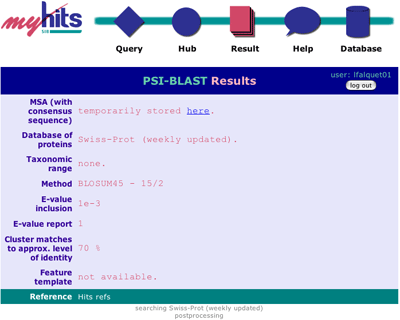 |
The top part summarizes your input data and options. Then you will see small messages about the evolution of the search. |
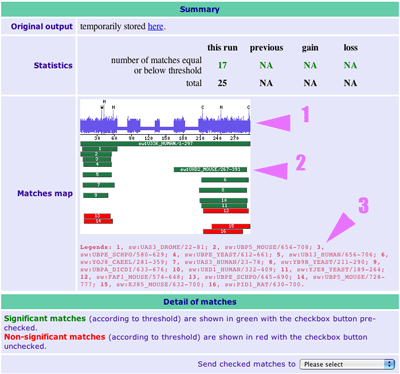 |
When the search is finished, you get the results in 3 parts. a) the statistics (useful to observe the evolution of the collected proteins) b) the graphic (described here) 1. Score per residue, with highly conserved ones shown in one-letter code 2. Position and significance of the matches 3. Description of the matches c) the alignments (see below) |
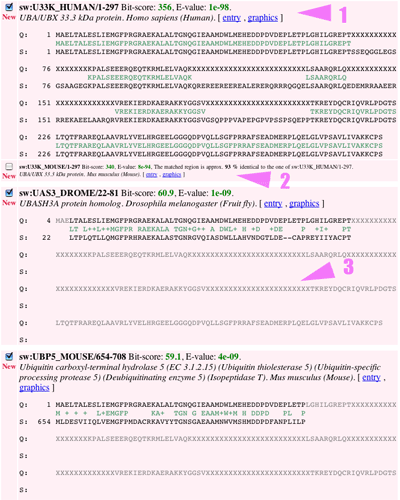 |
c) the alignments 1. Detailed description of the match (position, species, scores, alignment) with links to the original entry and to a graphical view 2. In a smaller font, a member of the same cluster (identity level above threshold) 3. In grey non-matching alignment |
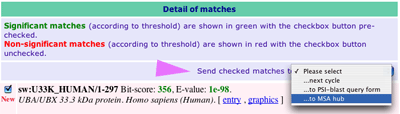 |
Check only the matches you want to keep (e.g., those matching the first 60 amino acids) and then use the drop down menu to send the matches either: - ...next cycle (redo a PSI-BLAST cycle with new PSSM from selected matches) - ...to PSI-BLAST form - ...to MSA Hub (please choose this option) |
| How to use the MSA Hub? | |
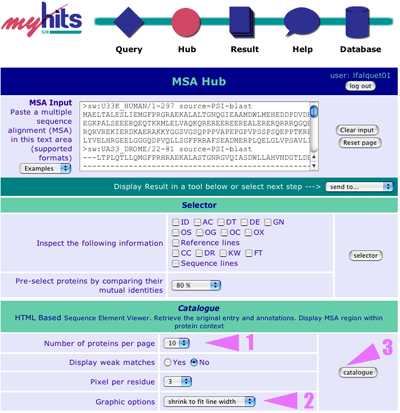 |
Here you can observe your multiple sequence alignment, modify it, and send it to another page. For example you want to use the "Catalogue". 1. Select "HTML walker [10 prot./page]" (10 proteins per page) 2. Select "shrink to fit line width" 3. Click on "Catalogue" button |
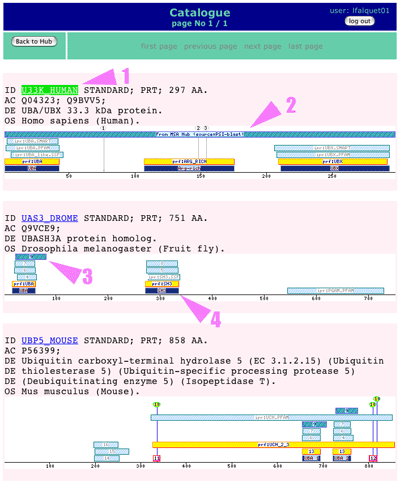 |
Now you can see one image for each matching protein with the description at the top: 1. Link to original protein entry 2. Shaded box displaying the position of the match 3. Same but for another protein 4. Other features found in the protein entry Now please click on the "back to Hub" button (top of the page), then in the MSA Hub click on the "jalview2" button |
| How to use Jalview? | |
 |
Please click again on the "Start Jalview" button to launch the Java applet in another window |
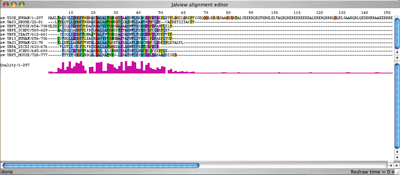 |
This is the view of the multiple alignment given by the Jalview applet. You can immediately notice that the matching region is only located in the first 60 amino acids. Using Jalview, please reduce the size of the alignment. (see Jalview documentation) |
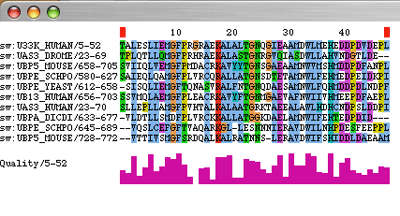 |
This is the reduced multiple sequence alignment. Once you are satisfied with it, please select the Jalview file sub-menu "Output to Browser" |
| What next? | |
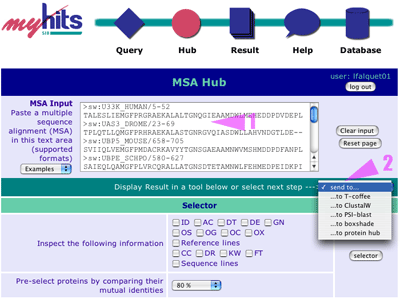 |
1. The modified multiple sequence alignment is now back in the text area 2. Select the "...to PSI-BLAST" to send your alignment to the PSI-BLAST form Comment: this alignment is very small and doesn't contain many gaps, thus you don't need to re-align the sequences. In the case of a longer and more complex alignment, we recommend using MAFFT to re-align the sequences before going back to PSI-BLAST. |
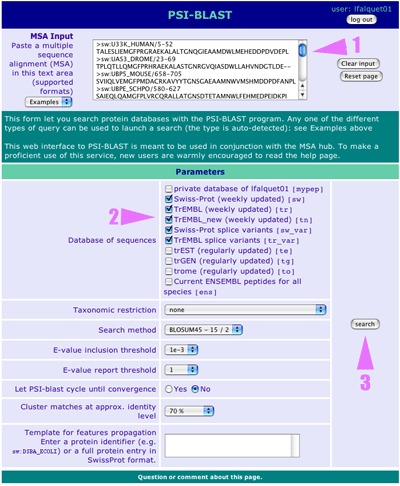 |
1. The modified multiple sequence alignment is now in the text area 2. Check other databases. (Note that the BLOSUM45 is magically kept from your first search) 3. Click on the search button to submit your multiple alignment to PSI-BLAST |
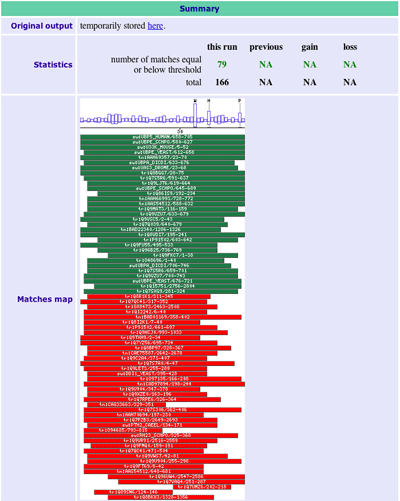 |
Congratulations you have defined a protein domain, and now you get more matches!! |